来源:BioArt
点评 | 何川(芝加哥大学)
撰文 | 小柚
作为细菌中丰度最高的DNA修饰,6mA直到2015年才首次被报道存在于真核生物的基因组中【1-3】。随后,许多研究的跟进发现6mA也存在于哺乳动物和高等植物的基因组。然而,这些研究对于6mA的丰度,分布和功能阐述有较大差异,并且由于6mA在基因组中的丰度较低,且其鉴定易受细菌污染的影响,目前关于6mA是否存在于真核生物基因组中,以及是否具有生理功能,还存在较大的争议(详见BioArt报道:学术争鸣 | 从迷雾重重到柳暗花明?全面起底DNA 6mA修饰相关研究的争议)。
2016年,耶鲁大学的Andrew Z. Xiao教授首次在小鼠胚胎干细胞中鉴定了DNA 6mA的存在【4】,然而另两个独立课题组却未能重复出相似的结果【5-6】,因此关于6mA在小鼠胚胎细胞中的鉴定和功能还需进一步研究。同时,何川教授等发现核基因组的6mA含量稀少,而线粒体基因组中的6mA则丰度更高【7】,提示线粒体是研究6mA功能的潜在理想模型,那么核基因组中的6mA是否还能获得研究者的关注呢?
2020年7月15日,Andrew Z. Xiao教授和清华大学的李海涛教授合作在Nature发表研究“N6-methyladenine in DNA antagonizes SATB1 in early development”,阐述了DNA 6mA修饰通过抑制SATB1在SIDD区的结合,从而调控染色质状态并影响胚胎早期发育进程的机制。

由于6mA在小鼠胚胎干细胞(mES)中的丰度较低(百万分之6-7),研究者首先寻找适宜的细胞培养条件来提高6mA的含量。值得注意的是,在四倍体补偿实验(tetraploid complementation,4N)中,6mA丰度与不同培养条件下mES的发育潜力呈正比。也就是说,在传统的2i(加入ERF和GSK3b抑制剂)条件中(4N阴性),6mA水平显著降低;而在其他2i条件下(4N阳性),6mA的水平则得以保留。mES细胞培养条件的差异可能是造成不同课题组关于6mA丰度争议的原因。
以小鼠滋养层干细胞(trophoblast stem cells)为模型,m6A的丰度在TS-like细胞时期显著上升,随后下降(图1 )。

图1
DIP-seq显示6mA主要位于AT含量高的基因间区域(intergenic regions),比如转座子LINE-1s,与之前的研究一致【4】。特别地,进一步分析显示6mA主要位于SSID区(stress-induced DNA double helix destabilization)。SSID区有助于推动拓扑压力诱导的DNA双螺旋不稳定,对染色质结构的组织有重要作用,包括建立和维持异染色质-常染色质边界和DNA的长距离互作等。SATB1是SSID的调节蛋白,可以直接结合并稳定DNA双螺旋结构。
那么,6mA是否会影响调节蛋白,比如SATB1,在SSID区的结合呢?确实,分析显示在in vitro和in vivo条件下,6mA都能显著的阻碍SATB1对DNA的结合。过表达6mA去甲基化酶ALKBH1后(注意,免疫荧光显示过表达的ALKBH1主要定位在细胞核,而非线粒体,可排除ALKBH1对线粒体DNA 6mA的影响),6mA降低的位点的染色质变得更加开放,异染色质区域也被打开,这说明6mA通过拮抗SATB1维持染色质的常染色质-异染色质边界。
为进一步研究6mA在胚胎发育中的功能,研究者构建了ALKBH1缺失的小鼠。虽然ALKBH1缺失的杂合小鼠可以出生,但是纯和小鼠却不能。ALKBH1的缺失抑制了TSC细胞向TGC细胞的分化,这说明6mA在胚胎发育中具有重要作用。同时,SATB1的敲除也有与ALKBH1缺失相似的表型,说明6mA主要通过拮抗SATB1发挥功能。
总的来说,该研究发现6mA通过拮抗SATB1在发育的早期调节染色质结构(图2),为研究表观遗传修饰调控染色质结构和基因表达提供了新的思路,具有重要意义。
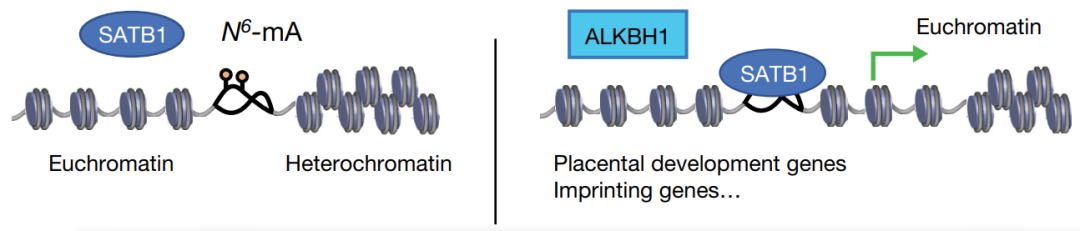
图2
专家点评DNA 6mA修饰在细菌界非常普遍,也存在于低等真核生物中,并已证明会影响转录。近年来,6mA被报道存在于哺乳动物基因组DNA中并具有潜在的功能。然而,由于它在细胞内的低丰度以及实验试剂中极易带来的细菌DNA污染, 目前关于gDNA中6mA是否存在正充满争议。我一直相信,正如我们以前在有关该主题的评论【8】中所写:
i)5mC是哺乳动物gDNA中占主导地位的DNA甲基化,发挥最主要的调节作用;
ii)6mA不太可能在哺乳动物gDNA上作为一种常见的表观遗传调控标记;
iii)6mA很有可能出现在5mC缺失的区域或发育时期,也可能在特定的压力反应或刺激下存在。从进化上讲,自然界利用细菌和低等真核生物中现有的腺苷甲基化来增加基因调节的复杂度是合理的。
我们确实需要承认,由于对6mA的潜在污染源缺乏了解,以及一些草率而且不负责任的研究,6mA的研究受到了挫折。最近进行的哺乳动物gDNA中6mA的仔细分析,对于确定正确的研究路线非常重要。考虑到这些因素,已有研究显示出哺乳动物线粒体DNA中显著存在6mA【7】,我们和其他人未发表的结果表明,哺乳动物细胞gDNA的 6mA在压力条件下会增加。
现在,Andrew Z. Xiao和李海涛实验室及其合作者进行了一项非常出色的研究,报告了在gDNA的SIDD区(stress-induced DNA double helix destabilization)存在6mA。作者显示出有力的证据支持在小鼠滋养细胞干细胞的这些区域中存在6mA。他们进一步表明,SATB1是一种与SIDD相互作用的蛋白质,在有或没有6mA的情况下,SATB1结合DNA的能力表现出500倍的差异。因此,作者提出了在胚胎早期发育过程中6mA调控染色质结构的功能。与5mC不同,6mA的存在使Watson-Crick碱基配对不稳定。DNA重复元件处6mA的存在很可能是使dsDNA不稳定的机制。从dsDNA到ssDNA的转变或dsDNA和ssDNA之间的平衡也被认为是基因表达调控的一种方式。6mA可能恰好适合此角色。
要提供充分令人信服的证据证明6mA的功能,仍然存在许多挑战。最主要的挑战是两个:i)鉴定介导gDNA 6mA甲基化的甲基转移酶;ii)一种真正的定量测序方法,可以鉴定出6mA修饰的准确位点和丰度。METTL4可以介导线粒体DNA(mtDNA)甲基化。mtDNA和gDNA可能存在其他甲基转移酶。真实的定量方法将打开整个领域。
附何川教授的点评原文:
DNA 6mA modification is prevalent in bacterial world. It is also present in low eukaryotes and have been shown to impact transcription. Its presence and potential functional roles in mammalian genomic DNA have been reported in recent years. Because of its low abundance and contaminations from bacterial DNA inside cells as well as in reagents, the presence of 6mA in gDNA has been challenged. I have always believed as we wrote in reviews (https://www.nature.com/articles/nsmb.3412 ) on this subject previously:
i) 5mC is the predominate DNA methylation that exerts most dominant regulatory roles on mammalian gDNA;
ii) the presence of 6mA as a common regulatory mark on mammalian gDNA is less likely;
iii) it is very possible that 6mA is present in regions or at developmental stages 5mC is depleted or not present. 6mA may also be present during specific stress response or under stimulation.
Evolutionally, it makes sense that nature would take advantage of the existing adenosine methylation in bacteria and low eukaryotes for adding additional layers of regulation complexity.
We do need to acknowledge that studies on 6mA have been set back by lack of knowledge about its potential source of contamination and sometimes just sloppy, irresponsible reports. Recent careful analysis of 6mA in mammalian gDNA is very important to bring cautions to the community and set the right course. With these cautions in mind, results have also been emerging to show notable presence of 6mA in mitochondrial DNA in mammals (https://www.cell.com/molecular-cell/pdfExtended/S1097-2765(20)30111-8), unpublished results indicating increased and detectable gDNA 6mA in mammalian cells under stress from us and other, and now the very nice study by Xiao and Li laboratories and their collaborators reporting the presence of 6mA at regions of SIDD. The authors showed strong evidence supporting the presence of 6mA at these regions in mouse trophoblast stem cells. They have further shown that SATB1, a protein interacts with SIDD, exhibits 500-fold difference in DNA binding with or without 6mA. The authors thus propose functional roles of 6mA-mediated regulation in chromatin domain separation during early embryo development. Different from 5mC, the presence of 6mA destabilizes Watson-Crick base pairing. The presence of 6mA at DNA repeat elements is very likely a mechanism to destabilize dsDNA. Transition from dsDNA to ssDNA or equilibrium between dsDNA and ssDNA have been proposed as another layer of genome regulation. 6mA could uniquely fit this role.
Many challenges remain exist to provide fully convincing evidence that 6mA is doing what has been proposed. On top of the lists are two main challenges: i) identification of the methyltransferase that mediates gDNA 6mA methylation; ii) a truly quantitative sequencing method that can convincingly read out the exact local and modification level at each 6mA modified site. METTL4 can mediate mitochondrial DNA methylation. Presence of other methyltransferases for mtDNA and gDNA is possible. A truly quantitative method will really open up the entire field.
来源:BioGossip BioArt
原文链接:https://mp.weixin.qq.com/s?__biz=MzA3MzQyNjY1MQ==&mid=2652491840&idx=1&sn=9b3700b2fae741eb50f78d46864e7290&chksm=84e243f4b395cae2ff566d671295211b06363ae6a523dc4b0565114caa99e52be26cffb0c747#rd
版权声明:除非特别注明,本站所载内容来源于互联网、微信公众号等公开渠道,不代表本站观点,仅供参考、交流、公益传播之目的。转载的稿件版权归原作者或机构所有,如有侵权,请联系删除。
电话:(010)86409582
邮箱:kejie@scimall.org.cn


